📗 -> 02/11/25: ECS124-L10
🎤 Vocab
❗ Unit and Larger Context
- Alignment
- BLAST
- Reference genome search - i.e. suffix tree
- Genome assembly
- Started covering De Novo assembly (no reference genome)
- Cycles
- Eulers Path
- Hamiltonian path
✒️ -> Scratch Notes
Sanger Sequencing via Scaffolding
Hamiltonian cycle instead reads - use k-mers of length 3: path that visits each node only once returns to the starts
| R1 | ATGGCGT | K-mers | Connections with K-mers[1:] | ||
|---|---|---|---|---|---|
| R2 | GGCGTGC | ATG | -> | TGG, TGC | |
| 3 | CGTGCAA | TGG | -> | GGC | |
| 4 | TGCAATG | GGC | -> | GCG | |
| 5 | CAATGGC | GCG | -> | CGT | |
| CGT | -> | GTG | |||
| GTG | -> | ||||
| TGC | -> | ||||
| GCA | -> | ||||
| CAA | -> | ||||
| AAT | -> |
- Can visualize as a big circle of connected nodes.
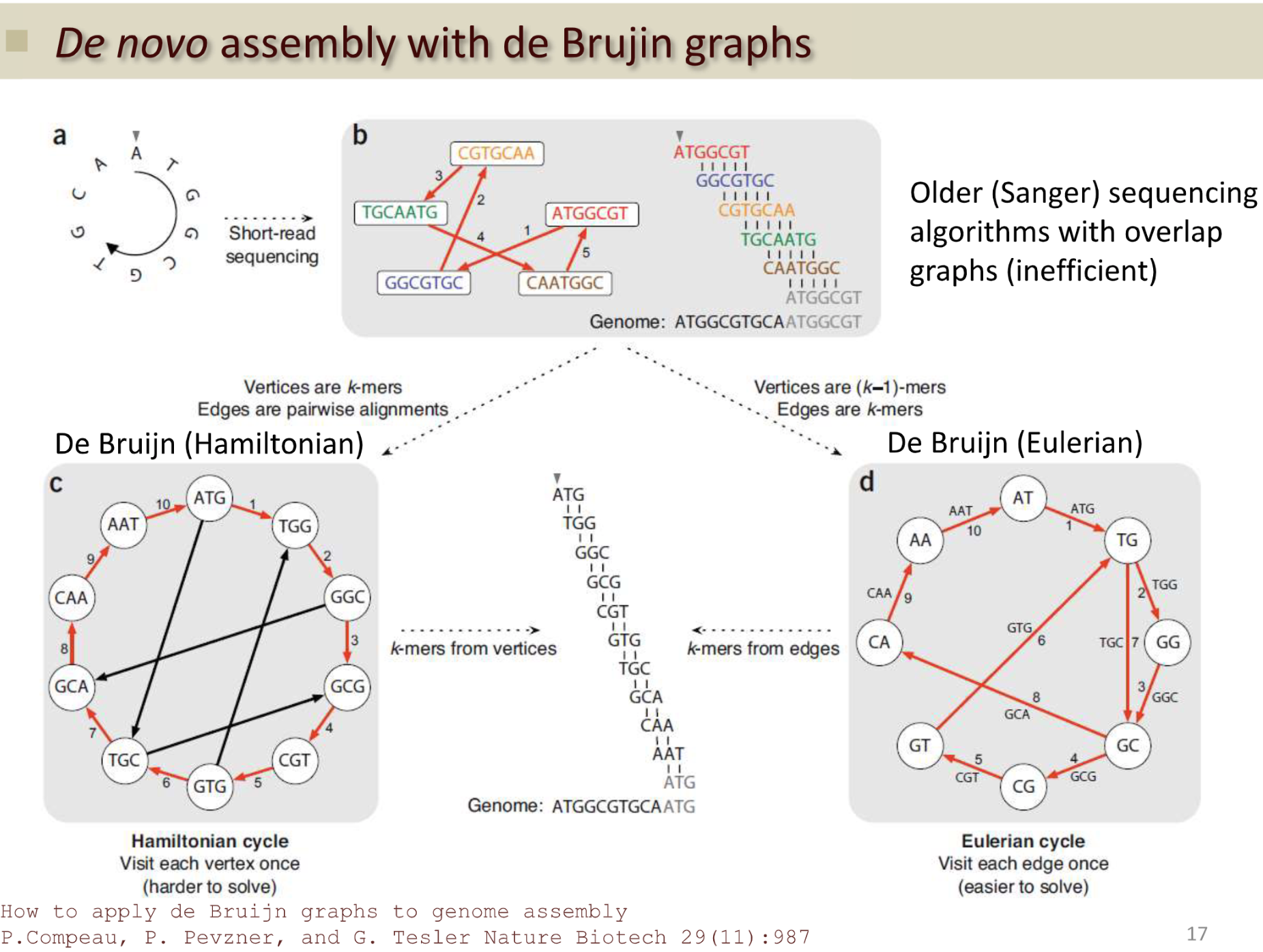
An Euler path is a path that passes through every edge exactly once. If it ends at the initial vertex then it is an Euler cycle.
A Hamiltonian path is a path that passes through every vertex exactly once (NOT every edge). If it ends at the initial vertex then it is a Hamiltonian cycle.
AT -> ATG -> TG
- Alignment
…
Machine Learning / Classification
Classify a region as coding or non coding, and annotate it in some way
Starting with Naive Bayes
Patient tests high protein.
| .8 | .25 | |||
|---|---|---|---|---|
| .2 | .75 | |||
blah blah rehash of ECS-132-Main
🧪 -> Refresh the Info
Did you generally find the overall content understandable or compelling or relevant or not, and why, or which aspects of the reading were most novel or challenging for you and which aspects were most familiar or straightforward?)
Did a specific aspect of the reading raise questions for you or relate to other ideas and findings you’ve encountered, or are there other related issues you wish had been covered?)
🔗 -> Links
Resources
- Put useful links here
Connections
- Link all related words